 |
On the taxonomy of genus Avena |
| Dr. Igor Loskutov |
 |
The global VIR oat collection is represented by comprehensive specific and intra-specific diversity of both cultivated (12,000 accessions) and wild (2,000 accessions) species of Avena. Full botanic and ecological diversity of cultivated species is incorporated in the landraces varieties-populations collected in 1910s - 1920s. A majority of these forms came from the centres of origin and diversity of this crop, providing a universal overview on the total geographic diversity of oat. With this in view, oat species became the subject of complex investigation in order to specify the system of the Avena genus, direction of its evolution and phylogenetic links between the species. At the same time, further search for taxonomy and utilization of new oat breeding sources for breeding purposes is one of the objectives pursued by Vavilov Institute of Plant Industry (VIR) in studying its global germplasm collections.
Based on the recent literature concerning the oats and on the evaluation of a representative set of oat accessions of 26 species VIR world collection of genus Avena L. under field conditions and different laboratory methods were confirmed that the genus can be divided up into two subgenus: Avenastrum C. Koch and Avena. Perennial autotetraploid A. macrostachya Balan. belongs to subgenus Avenastrum and other wild and cultivated species belong to subgenus Avena.
It is the availability of total botanical and eco-geographic diversity and its complex study that may provide an opportunity to identify centres of origin and variability of this or that genus or species.
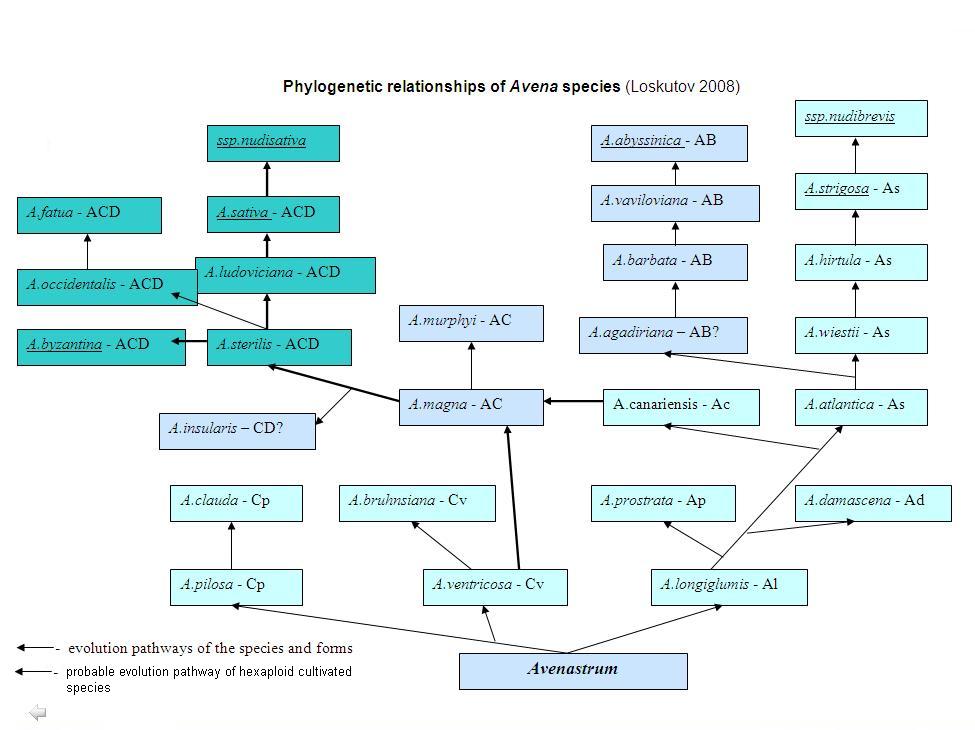
Complex analysis of broad literary review and utilization of the karyotype structure data confirmed by the results of RAPD and avenin spectrum analysis confirmed identification of two basic genomes which had most likely participated in the formation of species in Avena, namely the A and C genomes. As for the B and D genomes, they seem to be derivatives of the A genome. In addition to the data obtained during the study of the species containing these genomes, clear-cut differences have been discovered in the areas of their distribution.
Numerous researches have proven that the C genome goes through all ploidy levels unchanged, with small variation, thus being considered one of the basic genomes in oat. Our investigations of the karyotype A. macrostachya Balan. have shown that this species is an autotetraploid with the AA genome [Loskutov & Abramova, 1999]. On the other hand, the analysis of chromosome structure indicates that A. macrostachya is related to the C-genome species [Rodionov et al., 2005]. At the same time, this species is characterized by a symmetrical karyotype and a set of morphological characters attesting to its true primitivity. All this is confirmed by the most primitive perennial type of development and by cross-pollination, which is typical for a group of species of oat-like grasses within the subgenus Avenastrum C. Koch [Loskutov, 2003]. Meanwhile, according to A. V. Rodionov et al. [2005], the division of the phylogenetic oat lines carrying A and C genomes was accompanied by accumulation of differences in dispersed repetitions and accumulation of transitions and transversions specific for each branch. Later the C-genome line segregated phylogenetic branches of A. macrostachya (genome CmCm) from the progenitor of the other species with the C genome, and after that the progenitors of A. macrostachya doubled their chromosome number and generated large blocks of C-heterochromatin which caused an unusual C-banding pattern of chromosomes in C genomes of diploid and polyploid species.
Afterwards the A genome developed independently from the C genome, which brought about lots of A-genome variants (Al, Ap, Ad, Ac, As), and finally produced a cultivated diploid species (A. strigosa Schreb.) with the As genome. RAPD analysis and studying avenin protein markers made it possible to conclude that in spite of all differences between species with the A genome they have indirect evolutionary affinity [Loskutov et al., 1999; Loskutov & Perchuk, 2000]. Genesis of tetraploid species became possible either after the doubling of the chromosome number in one of the diploid species (AA) or with spontaneous hybridization of two closely related (AB = AA') diploid species. This resulted in raising ploidy to a higher level and bringing into existence a group of tetraploid species with either AB or AA' genomes, which provided an opportunity for the development of a cultivated tetraploid species (A. abyssinica) containing the AB genome. Later, diploid species with A and C genomes united into one genotype (A. canariensis, Ac and A. ventricosa, Cv), where the A genome in one of intermediate forms transformed by structural divergence into a D genome or, as it is now assumed, into an A" genome. The species with A and AB (AA') genomes and a biaristulate lemma tip (sectio Aristulatae (Malz.)) had in most cases a disarticulated floret. Some of them have cultivated analogues with the same ploidy level (A. wiestii Steud., A. hirtula Lagas. - A. strigosa Schreb.; A. vaviloviana (Malz.) Mordv.- A. abyssinica Hochst.) and wider areas of distribution (A. wiestii, A. hirtula and A. barbata), being rather active weeds (A. clauda Dur., A. pilosa M.B., A. damascena Rajh. et Baum, A. longiglumis Dur. and A. barbata). Obviously, this group seems apparently had no part in the development of hexaploid oats [Loskutov, 2003].
This publication presents historical review of botanical systems of genus Avena. Modern taxonomy is presented on the basis of complex evaluation of Avena species. It has been shown that the species with the C and AC genomes, whose characteristic feature, i.e. the presence of a bidentate lemma tip (section Avenae), is typical for hexaploid species, are transitional ancestral forms (looks like A. ventricosa, A. canariensis or A. magna) in the evolution of hexaploid oats. This group includes diploid species A. ventricosa Balan., A. bruhnsiana Grun., A. canariensis Baum and A. agadiriana Baum et Fed. as well as tetraploid species A. magna Murph. et Terr., A. murphyi Ladiz. and A. insularis Ladiz., that bear only disarticulated spikelets and do not have direct cultivated analogues.
Significant differences between tetraploid species with AB and AC genomes have been confirmed by the data of RAPD analysis and avenine protein markers [Loskutov et al., 1999; Loskutov & Perchuk, 2000]. Further on, the species with three genomes A, C and D underwent hybridization and produced an allohexaploid species, the progenitor of A. sterilis, which generated a large group of species, including hexaploid A. byzantina and A. sativa with the ACD (ACA") genome. Divergence of A and C, two major genomes of the genus Avena, may be traced by the karyotype structure, avenine protein marker spectra and RAPD data [Loskutov & Abramova, 1999; Loskutov et al., 1999; Loskutov & Perchuk, 2000]. Besides, distinctive differences were found in the areas of distribution of the species containing these genomes [Loskutov, 2003].
Acting as such transitional progenitors for cultivated hexaploid species of Avena, in our opinion, may be wild diploid and tetraploid forms possessing a characteristic feature typical for hexaploids, that is the presence of two denticles on the tip of the lemma (section Avenae Losk.).
Presumably in the western part of the Mediterranean region, where the richest specific diversity of Avena is concentrated, spontaneous hybridization of diploid and tetraploid species from the group of transitional forms with genomes A, C and D initiated development of all allohexaploid species. The occurrence of the largest diversity of polyploids in the eastern part of Anterior Asia, where soil and climate conditions are harder than in the western Mediterranean areas, was confirmed by N. I. Vavilov's [1926] statement about greater hardiness of this group of species, as compared with diploid ones, because allopolyploid species promote development of extremely differentiated ecotypes, which played an important role in the evolution. Proceeding from the centre of origin toward the South-Western Asiatic centre, smaller-seeded and more adaptive hexaploid forms of wild species began to occur.
 |
Speciation in the genus Avena L. (Loskutov, 2007; Loskutov & Rines, 2011)
| Section | Species | 2n | Genome | |||
| Wild | cultivated | |||||
| floret disarticulation | spikelet disarticulation | |||||
| Aristulatae (Malz.) | A.clauda Dur. | A.pilosa M.B. | 14 | Cp | ||
| Losk. | A.prostrata Ladiz. | Ap | ||||
| A.damascena Raj.et Baum | Ad | |||||
| A. longiglumis Durie. | Al | |||||
| A. wiestii Steud. | A. atlantica Baum | As | ||||
| A. hirtula Lagas. | A. strigosa Schreb. | As | ||||
| A. barbata Pott. | 28 | AB | ||||
| A. vaviloviana Mordv. | A. abyssinica Hochst. | AB | ||||
| Avenae Losk. | A. ventricosa Bal. | 14 | Cv | |||
| A. bruhnsiana Grun. | Cv | |||||
| A. canariensis Baum | Ac | |||||
| A. agadiriana Baum et Fed. | 28 | AB? | ||||
| A. magna Mur. et Terr. | AC | |||||
| A. murphyi Ladiz. | AC | |||||
| A. insularis Ladiz. | AC? | |||||
| A. fatua L. | A. sterilis L. | A. byzantina C. Koch | 42 | ACD | ||
| A. occidentalis Durie. | A. ludoviciana Durie. | A. sativa L. | ACD | |||
|
|
|
| 1. Mediterranean centre - Morocco, Algeria, Spain | 5. South-West Asian centre - Turkey, Iran, Iraq, Syria |
| 2. Spain and Portugal - centre of diversity of A. strigosa | 6. Tatarstan, Bashkortostan - diversity of A. sativa convar. volgensis |
| 3. Great Britain - centre of diversity of A. strigosa subsp. nudibrevis | 7. China, Mongolia - centre of diversity of A. sativa subsp. nudisativa |
| 4. Abyssinian centre - Ethiopia, centre of diversity of A. abyssinica | -> - pathways of distribution of cultivated species and forms. |
|
 |